2. Theoretical principles:
2.1. Sequence Alignment:
OMEGAbase uses programs that take as input nucleotide sequences, build a multiple sequence alignment (using Tcoffee a Meta-Multiple Sequence Alignment Tool) of proteins and then transforms it back to DNA, into a codon alignment. The server automatically assigns the corresponding codon sequence even if the input DNA sequence has mismatches with the input protein sequence. Therefore, OMEGAbase can deal with in-frame stop codons and/or frameshift disruptions in the input alignment, which is suitable for the analysis of pseudogenes.
2.2. Selective pressure acting on genes:
Definition: The dN/dS ratio (or ω), is the ratio of the rate of non-synonymous substitutions (dN) to the rate of synonymous substitutions (dS), which can be used as an indicator of selective pressure acting on a protein-coding gene.
Methods: Methods for estimating dN and dS are classified into two groups: approximate methods and maximum-likelihood methods.
Approximate methods involve three basic steps:
- counting the number of synonymous and nonsynonymous sites
- counting the number of synonymous and nonsynonymous substitutions
- correcting for multiple substitutions
The maximum-likelihood approach uses the probability theory to combine all three steps in one step.
→ OMEGAbase uses the Codeml program from the computer package PAML. It estimates various parameters (Ts/Tv, ω = dN/dS, branch length) on the codon (nucleotide) alignment, according to the phylogenetic tree. The parameter of interest to the server is ω, which quantifies selective pressure acting on protein-coding region, and uses a maximum-likelihood method (Goldman and Yang 1994) to estimate different dN/dS values among branches and among sites.
• Maximum Likelihood Models:
The branch-site models allow to answer what proportion of sites in a lineage are under positive selection ? and in which lineage(s) have sites experienced positive selection?
OMEGAbase uses the branch-site model, which estimates different dN/dS values among branches and among sites. In this model, a branch of interest is selected and is called "foreground" branch. All other branches are called the "background" branches in the tree. The background branches share the same distribution of ω (dN/dS) value among sites, whereas different values can apply to the foreground branch.
In the branch-site analysis, two models and a Likelihood Ratio Test are computed (Zhang et al. 2005):
• A null model (H0), in which the foreground branch may have different proportions of sites under neutral selection than the background (i.e. relaxed purifying selection)
• An alternative model (H1), in which the foreground branch may have a proportion of sites under positive selection.
• A Likelihood Ratio Test (LRT) is computed with an associated pvalue.
→ The alternative model (H1):
There are three classes of sites on the foreground branch
ω0: dN/dS < 1
ω1: dN/dS = 1
ω2: dN/dS ≥ 1
All codon sites are then categorized into 4 classes 0, 1, 2a, and 2b with proportions of p0, p1, p2a, and p2b, respectively, as:
p0 : Proportion of sites that are under purifying selection (ω0 < 1) on both foreground and background branches.
p1 : Proportion of sites that are under neutral evolution (ω1 = 1) on both foreground and background branches.
p2a: Proportion of sites that are under positive selection (ω2>1) on the foreground branch and under purifying selection (ω0 < 1) on background branches.
p2b: Proportion of sites that are under positive selection (ω2>1) on the foreground branch and under neutral evolution (ω1 = 1) on background branches.
For each category, we get the proportion of sites and the associated dN/dS values.
→ The null model (H0) (ω2 fixed to 1):
p0 : Proportion of sites that are under purifying selection (ω0 < 1) on both foreground and background branches.
p1 : Proportion of sites that are under neutral evolution (ω1 = 1) on both foreground and background branches.
p2a: Proportion of sites that are under neutral evolution (ω2 = 1) on the foreground branch and under purifying selection (ω0 < 1) on background branches.
p2b: Proportion of sites that are under neutral evolution (ω2 = 1) on the foreground branch and under neutral evolution (ω1 = 1) on background branches.
→ The Likelihood Ratio Test (LRT)
For each model, we get the log likelihood value (lnL1 for the alternative model and lnL0 for the null model), from which we compute the Likelihood Ratio Test (LRT).
The 2x(lnL1-lnL0) follows a X² curve with degree of freedom o 1, so we can get a pvalue of the LRT.
Positive selection is inferred for the foreground branch if the LRT is greater than X² = 3.84 (5% significance level)
• Detect sites that have experienced positive selection
In cases where positive selection is inferred, the posterior probability of a site belonging to
the positively selected class is estimated using Bayes Empirical Bayes (BEB) calculation.
If BEB is predicted, positively selected sites with posterior probability cutoffs ≥0.95 are returned.
2.3. OMEGAbase pre-computed data sets:
Pre-computed values of selective pressure are stored in OMEGAbase for >100,000 genes for which we refined orthologuous relationships among 10 mammalian species (Human, Chimpanzee, Orang-utang, Macaque, Marmoset, Mouse, Rat, Dog, Cow, Horse). Results have been obtained with the codeml program from the PAML5 package (Branch-site test as set as model=2, NSsites=2).
Entries are:
• Selective pressure type
→ positive selection
→ negative/neutral selection
• Specificity of selection
→ lineage-specific positive selection
→ Shared (co-occurency) positive selection among lineages
• Proportion of sites under positive selection
→ Number of positive selected sites (codon)
→ Similarity or difference of positive selected sites between PSG among lineages
• Identification of sites under positive selection
→ Codon and position identification of positive selected sites (codon)
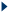 -home page- http://dogs.genouest.org/OMEGA
-home page- http://dogs.genouest.org/OMEGA
© Copyright 2005-2012 UMR6290-CNRS Last updated : July, 2012 Contact : christophe.hitte[at]univ-rennes1.fr
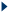 For batch query sequences : separate the set of orthologous sequences by '%'
For batch query sequences : separate the set of orthologous sequences by '%'